Sanger Method of DNA Sequencing-An Overview
The Sanger Method of DNA Sequencing, also known as dideoxy sequencing or chain termination sequencing uses fluorescently labeled dideoxy-nucleotides to terminate DNA synthesis at specific points, allowing researchers to determine the nucleotide sequence of DNA.
One of the key methods used in the Human Genome Project is Sanger Sequencing, and it is the best method for studying genes. It’s also the basis for many genetic tests today. Frederick Sanger and his team were the first to develop the technology that is now called DNA sequencing. It has been the most used method for about 40 years.
Sanger Method of DNA Sequencing
Sanger Sequencing was developed by Nobel Laureate Frederick Sanger and his colleagues in 1977. Since 1986, it has been commercially available thanks to Applied Biosystems, which invented it in 1985. The use of next-generation techniques for large-scale, automated, and/or automated genome analyses have replaced the use of high-throughput DNA Sequencing.
For validation of small-scale projects and the sequence of genomes, the Sanger method is still used. It is still cheaper than other methods for getting accurate, long-range DNA sequences, even though it does not have the advantage of long-read sequencing.
Automation And Sample Preparation
It is possible to sequence up to 384 DNA samples in a single piece of equipment. The machine that separates the strands of DNA is called a DNA scenographer. It uses capillary electrophoresis to record and report fluorescent dyes.
The data is outputted as peak traces. Some labeling reactions are performed separately and loaded on the scenographer. The peaks that are of a lower quality are discarded after each peak is scored. Large classes of anomalies in genes and other biological phenomena can be detected with denoising techniques. The new machine is not perfect, but it is accurate enough to detect many small variations of a given class of biological anomalies.
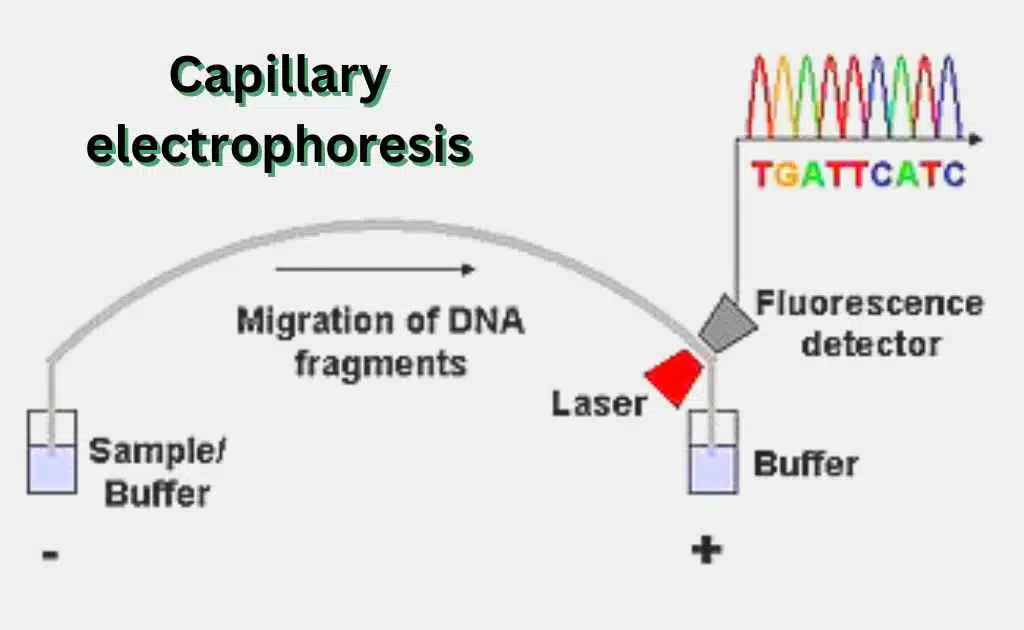
Process Of Sequencing
A DNA sample of interest is used as a template. When the sample DNA is combined with primer, DNA polymerase, and DNA nucleotides, it is added to a tube.
Each of the four dye-labeled, chain-terminating Dideoxynucleotides is added, but in much smaller amounts than the ordinary nucleotides.
The mixture is first heated to denature the template DNA, then cooled so that the primer can bind to the single-stranded template.
Once the primer has bound, the temperature is raised, and the polymerase enzyme synthesizes a new strand of DNA, starting from the primer.
A DNA sample of interest is used as a template. DNA polymerase will continue to add nucleotides to the chain until it adds a dideoxy nucleotide instead of a normal one.
If no further nucleotides can be added, the DNA strand will end with the dideoxy nucleotide.
There are many cycles in which this process repeats itself. By the time the process is completed, it’s virtually certain that dideoxy nucleotides will have been incorporated at every single position of the target DNA in at least one reaction.
A short DNA strand can be thought of as being made up of a series of bases arranged in a specific order. The ends of the strands will be tagged with special nucleotides, which will tell the scientists what kinds of nucleotides to insert at the ends of the fragment.
Detection of DNA Sequence
Each band of the gel, one by one, is read by a computer and the identity of each terminal nucleotide is determined. A laser can excite the fluorescent tags in each band, making it a perfect example of an excitation device. If a computer can detect light, that’s a perfect example of a detector.
There are different ways in which the dNTPs are labeled. Each of them emits its unique signal. The process by which cells replicate the genome is known as DNA replication. In this process, a new daughter cell gets a complete copy of its parent’s genome.
The purity of a chemical component is determined by the separation of mass, size, and other properties of the component. A chromatogram is a representation of the components separated by this technique.
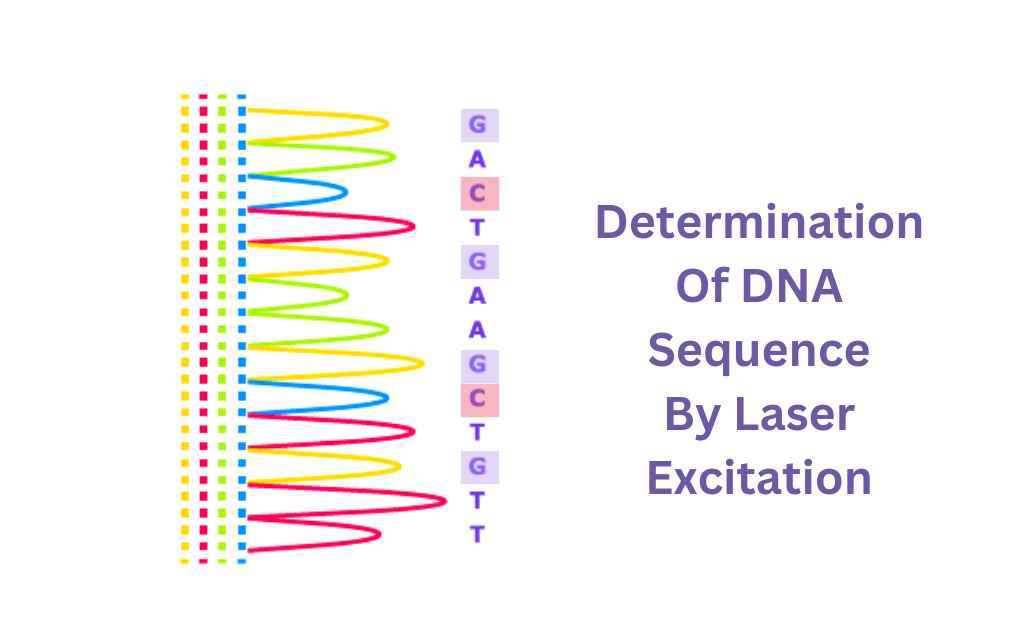
Latest Research About Sanger Method of DNA Sequencing
- A new technique called qSanger has been developed to detect neutral genetic mutations using mixed Sanger sequencing reads, providing a faster and more cost-effective alternative to traditional methods. Validated in vitro and in Escherichia coli, this method can identify single-base polymorphisms and de novo mutations with high accuracy. The qSanger method is poised to become a valuable tool for genetic research, offering a more efficient way to analyze DNA sequences and better understand the genetic basis of disease. [1]
- Scientists compared Sanger Method of DNA Sequencing and nanopore sequencing in identifying bacterial colonies from bioaerosol samples. Nanopore sequencing was superior in identifying individual bacterial components in multispecies colonies. [2]
- Scientists have developed a fast and reliable Sanger Method of DNA Sequencing protocol for identifying POLE exonuclease domain mutations in endometrial cancer tissues, which is still the most accurate and robust technique for sequencing DNA. [3]
- Scientists have developed a hands-on approach to teaching the Sanger Method of DNA Sequencing using paper clips to demonstrate chain termination. This method simplifies the chemical reaction of bond formation and helps students understand why dideoxy-nucleotides cause chain termination. [4]
Related FAQs
What are the three main steps of Sanger sequencing?
Selection of Sample
Size Separation By Gel Electrophoresis
Gel Analysis & Determination Of DNA Sequence
What is the purpose of the Sanger method?
Sequencing of a gene’s nucleotide sequence by Sanger technology is the method of choice for the analysis of many DNA segments.
What are the limitations of Sanger sequencing?
It only sequences short pieces. Short pieces of DNA between 300 and 1000 base pairs can only be sequenced by this method. In the first 15 to 40 bases, the quality of a Sanger sequence is not great. Sequence quality goes down after 700 to 900 bases.
What is the main enzyme used in Sanger sequencing?
The Klenow enzyme is a type of proteolytic fragment of the DNA polymerase enzyme, that is used in Sanger sequencing.
How many primers are used in Sanger?
Sanger sequencing uses only a single primer.



Leave a Reply